First, let’s make sure the anatomy of prokaryotes are familiar to us:
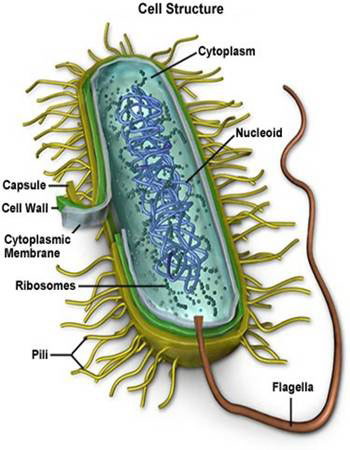 Prokaryotes are the simplest living cells, typically 1~10μm in diameter, and are found in all environmental niches from the guts of the animals to acidic hot springs.
Prokaryotes are the simplest living cells, typically 1~10μm in diameter, and are found in all environmental niches from the guts of the animals to acidic hot springs.
- They are bounded by a cell (plasma)membrane comprising a lipid bilayer, in which are embeded proteins that allow the exit and entry of small molecules.
- Most prokaryotes also have a rigid cell wall outside the plasma membrane, which prevents the cell from swelling or shrinking in environments where osmolarity differs significantly from that inside the cell.
- Sometimes the cell wall is surrounded by an (often) polysaccharide envelope called capsule.
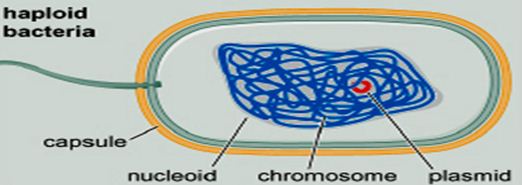
- The cell interior (cytoplasm or cytosol) usually contains a single, circular chromosome compacted into a nucleoid and attached to the membrane.
- There are no distinct subcelluar organelles in prokaryotes as in eukaryotes(except for the ribosomes核糖体).
- The surface of a prokaryote may carry pili, which allow the prokaryote to attach to other cells and surfaces. Some prokaryotes also carry flagella, whose rotating motion allows the cell to swim.
====================================================
To talk about prokaryotic chromosome structure, we use E. coli (大肠杆菌)as the example.
- E. coli chromosome contains a single supercoiled circular DNA molecule of length 4.6 million bp.
- E. coli chromosome is highly folded: 1500 µm of DNA length versus ~1 µm of cell size, forming a structure called the nucleoid.
- The nucleoid has a very high DNA concentration, perhaps 30~50 mg/ml, as well as containing all the proteins associated with DNA, such as polyerases, repressors(a protein that is determined by a regulatory gene, binds to a genetic operator, and inhibits the initiation of transcription of messenger RNA), etc.
 ——————————————————————————-
——————————————————————————-
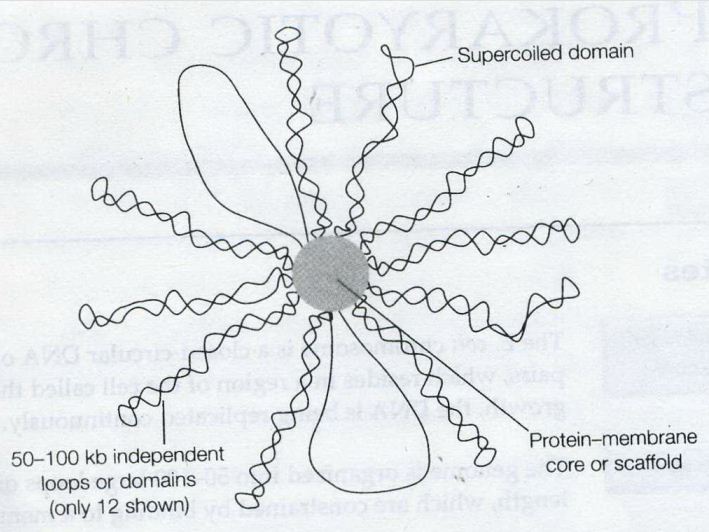
DNA domains (loops)
 Remember this famous electron micrograph of an E. coli cell we showed before? The cell was carefully lysed, all the proteins removed and then, it was spread on an EM grid to reveal all of its DNA.
Remember this famous electron micrograph of an E. coli cell we showed before? The cell was carefully lysed, all the proteins removed and then, it was spread on an EM grid to reveal all of its DNA.- The DNA consists of 50~100 domains or loops, about 50~100 kb in size (kb: kilobase, a unit of measure of the length of a nucleic-acid chain that equals one thousand base pairs).
- The ends of the loops are constrained by binding to a structure which probably consists of proteins attached to part of the cell membrane.
- Not known whether the loops are static or dynamic, but one model suggests that the DNA may spool(wind) through sites of polyerase or other enzymic action at the base of the loops.
Supercoiling of the genome
The E. coli chromosome as a whole is negatively supercoiled, although there is some evidence that indicates individual domains may be supercoiled independently. Electron micrographs indicate that some domains may not be supercoiled, perhaps because the DNA has become broken in one strand, where other domains clearly do contain supercoils.
The domains may be topologically independent. There is, however, no real biochemical evidence for major differences in the level of supercoiling in different regions of the chromosome in vivo
DNA-binding proteins
The looped DNA domains of the chromosome are constrained further by inter-action with a number of DNA-binding proteins.
The most abundant of these are protein HU, a small basic (+charged) dimeric protein, which binds DNA non-specifically by the wrapping of the DNA aorund the protein.
And H-NS (formerly known as the protein H1), a monomeric neutral protein, which also binds DNA non-specifically in terms of sequence, but seems to have a preference for regions of DNA which are intrinsically bent.
These proteins are sometimes known as histone-like proteins, and have the effect of compacting the DNA, which is essential for the packaging of the DNA into the nucleoid, and of stabilizing and constraining the supercoiling of the chromosome.
![the blueprint of life [7]: prokaryotic chromosome structure of DNA](https://apbiology.cn/wp-content/uploads/sites/8/2013/07/m-bio-7-bacteria.jpg)
