Finally, we come to the last part of the molecular structure of DNA.
4. Tertiary Structure of DNA – superhelix structure
Advanced folding and intertwining of DNA molecules over the secondary structure .
DNA topology
1. Linear DNA:
- Commonly seen in eukaryotes,
- with extreme length,
- complementary sequence
- included in the chromatin (the combination or complex of DNA and proteins that make up the contents of the nucleus of a cell)
- interacting with other cellular components.
2. Circular DNA:
(DNA frequently occurs in nature as closed-circular molecules, where the two single strands are each circular and linked together. The number of links is known as the linking number(Lk).)
- Usually seen in prokaryotes, e.g. plasmid (质粒), circular bacterial chromosomes and many viral DNA molecules
- cccDNA (covalently closed circular DNA) → supercoiled or Relaxed; ncDNA (nicked circular DNA): a nick (缺刻)formed by breaking one phosphodiester bond
- two complementary single strands are each joined into circles, 5′ to 3′, and are twisted around one another by the helical path of DNA.
- The molecule has no free ends and the two single strands are linked together a number of times corresponding to the number of double-helical turns in the molecule.
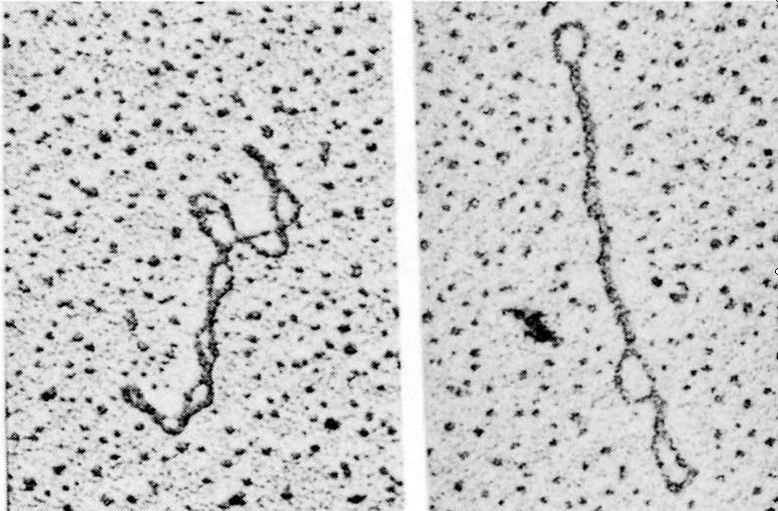
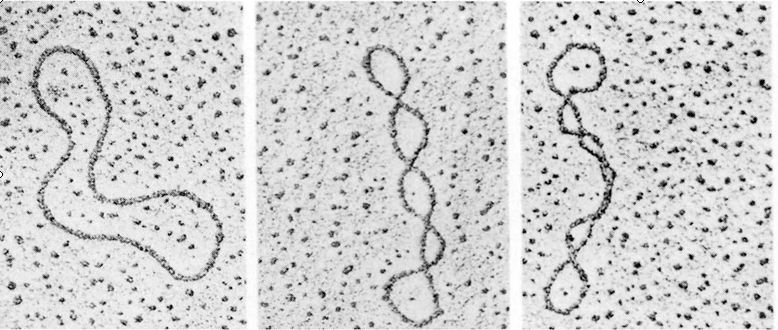
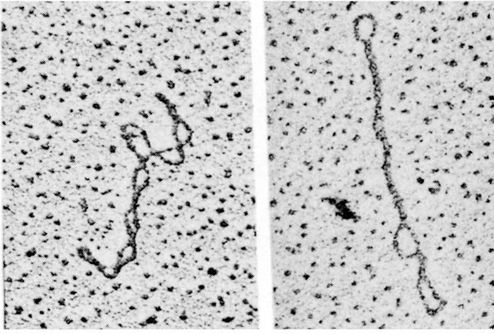
cccDNA Topology
- Supercoiled DNA
Negative superhelix: natural status of cccDNA with less intra- molecular tension (underwound effect)
Positive superhelix: unnatural status with higher tension (overwound effect)
- Relaxed circular DNA: intermediate between negative superhelix and positive superhelix
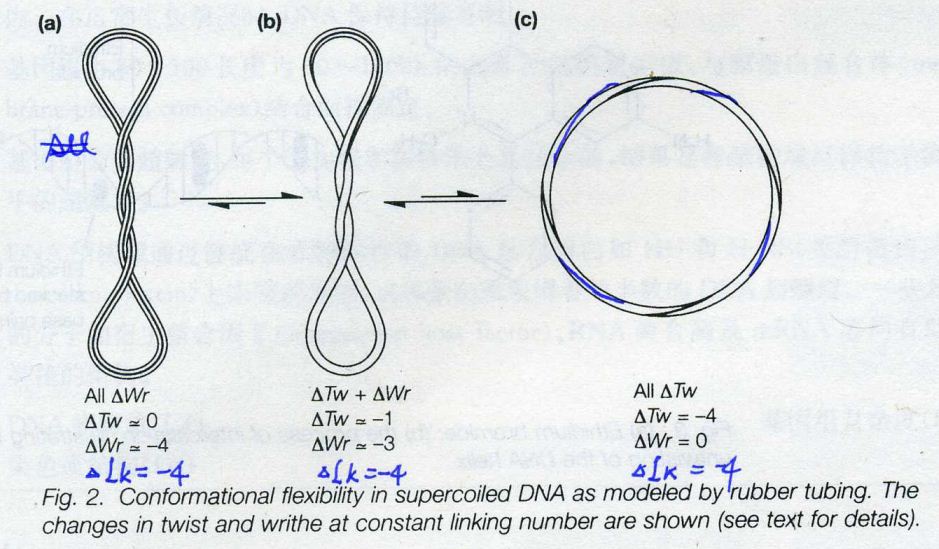
- Topological Equation of cccDNA
L = T + W
L=Linking number=total number of times one strand of the double helix links the other
T=Twisting number= the number of times one strand completely wraps around the other strand
W=Writhing number= the number of times that the long axis of the double helical DNA crosses over itself in 3-D space
Features
1. The linking number of a closed-circular DNA is a topological property, that is one which cannot be changed without breaking one or both of the DNA backbones. (A molecule of a given linking number is known as a topoisomer. Topoisomers differ only in their linking number)
2. Twisting number
For B DNA, T>0(10 bp per turn)
For A DNA, T>0(10.5 bp per turn)
For Z DNA, T<0 (12 bp per turn)
3. Writhing number
Relaxed: W=0
Negative supercoils: W<0
Positive supercoils: W >0
- Topoisomerases (enzymes used to regulate the level of supercoiling of DNA molecules)
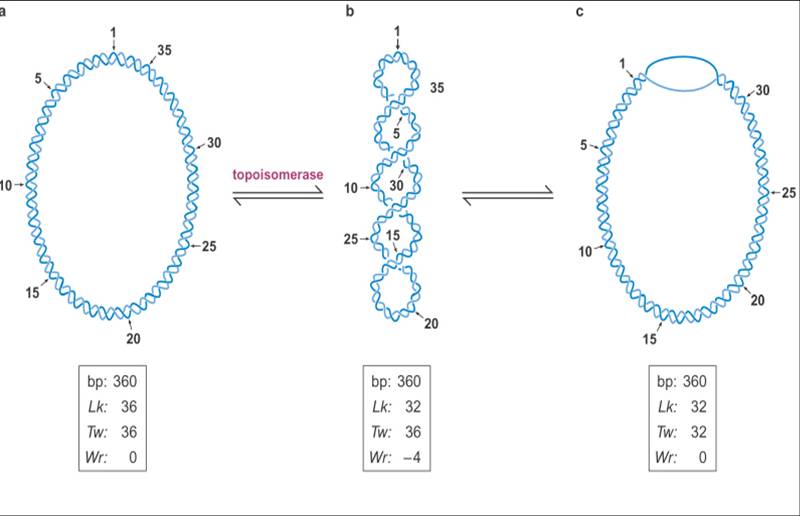
To alter the linking number of DNA, the enzymes must transiently break one or both stands. There are two classes of topoisomerases:
Type I Topoisomerase nick one strand of the DNA, changing 1 Linking-number at a time(+/-1 Lk)
Type II Topoisomerase, which requires the hydroloysis of ATP, break two strands of DNA, changing 2 Linking-number at a time(+/-2Lk); also able to unlink DNA molecules.
Most topoisomerases reduce the level of positive or negative supercoiling, that is, they operate in the energetically favorable direction. (However, DNA gyrase, a bacterial type II enzyme, uses the energy of ATP hydrolysis to introduce negative supercoiling into hence removing positive supercoiling generated during replication.)
Topoisomerases are essential enzymes in all organisms; they are involved in replication, recombination and transcription.
Both type I and II enzymes are the target of anti-tumor agents in humans.
DNA superhelix
Biological significance of Superhelix:
- DNA packing:
Eg.


This is a famous electron micrograph of an E. coli cell that has been carefully lysed, then all the proteins were removed, and it was spread on an EM grid to reveal all of its DNA.
- DNA functioning:
Negative supercoils serve as a store of free energy that aids in processes requiring strand separation, such as DNA replication and transcription; Strand separation can be accomplished more easily in negatively supercoiled DNA than in relaxed DNA.
1. DNA in cells is negatively supercoiled;
2.(-)supercoiling introduced by a topoisomerase II (gyrase 促旋酶) in prokaryotes and by nucleosome (核小体)in eukaryotes
3.Cruciform or bubble structures introduced by (-) supercoiling are potential protein-binding sites