Let’s pick up where we dropped, the secondary structure of DNA.
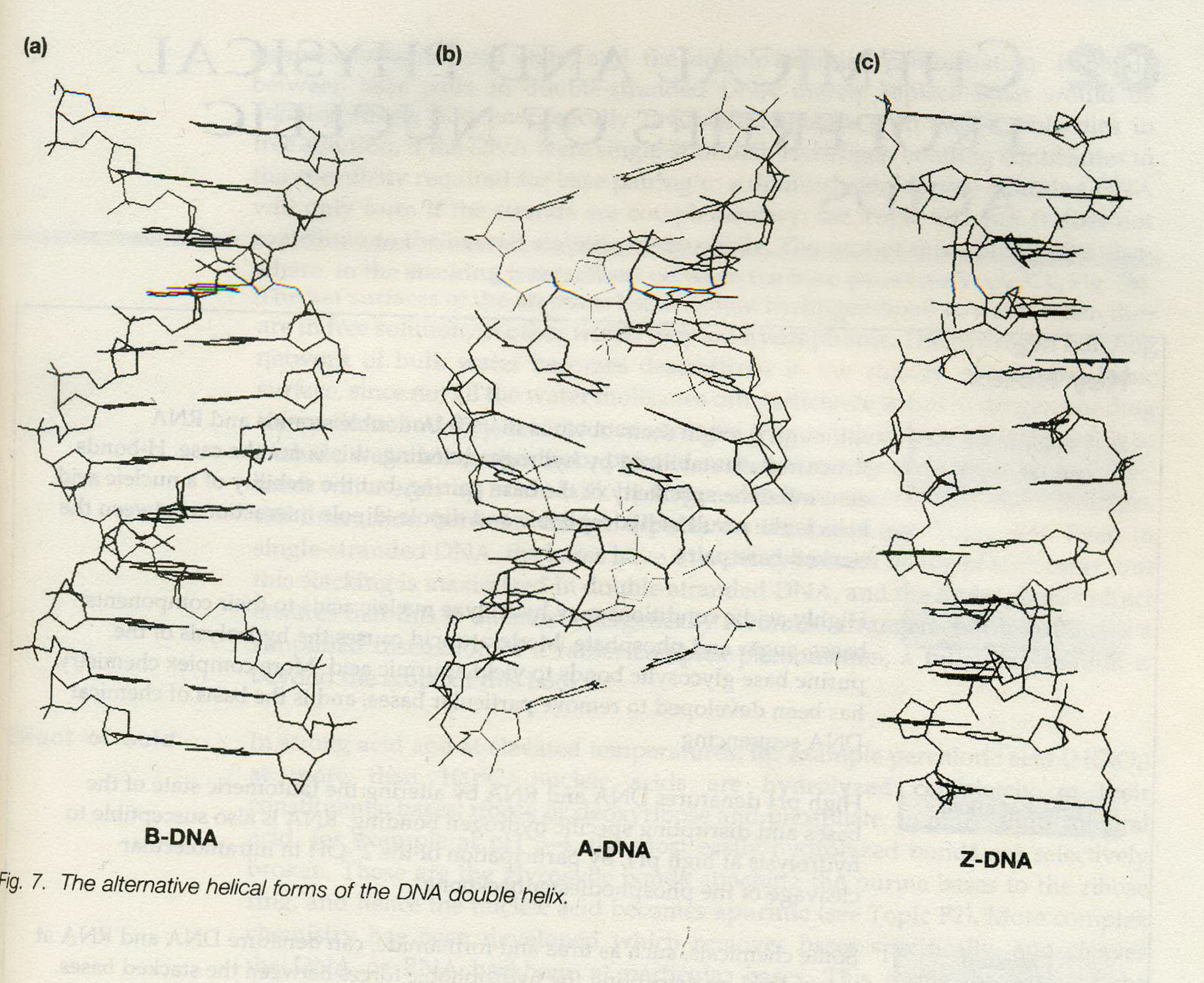
IN FACT, a number of different forms of nucleic acid double-helix have been observed and studied, all having the basic pettern of two helically-wound antiparallel strands.
Polymorphism (多样性)of DNA Secondary Structure
―due to conformational changes of sugar-ring on the nucleotide chain.
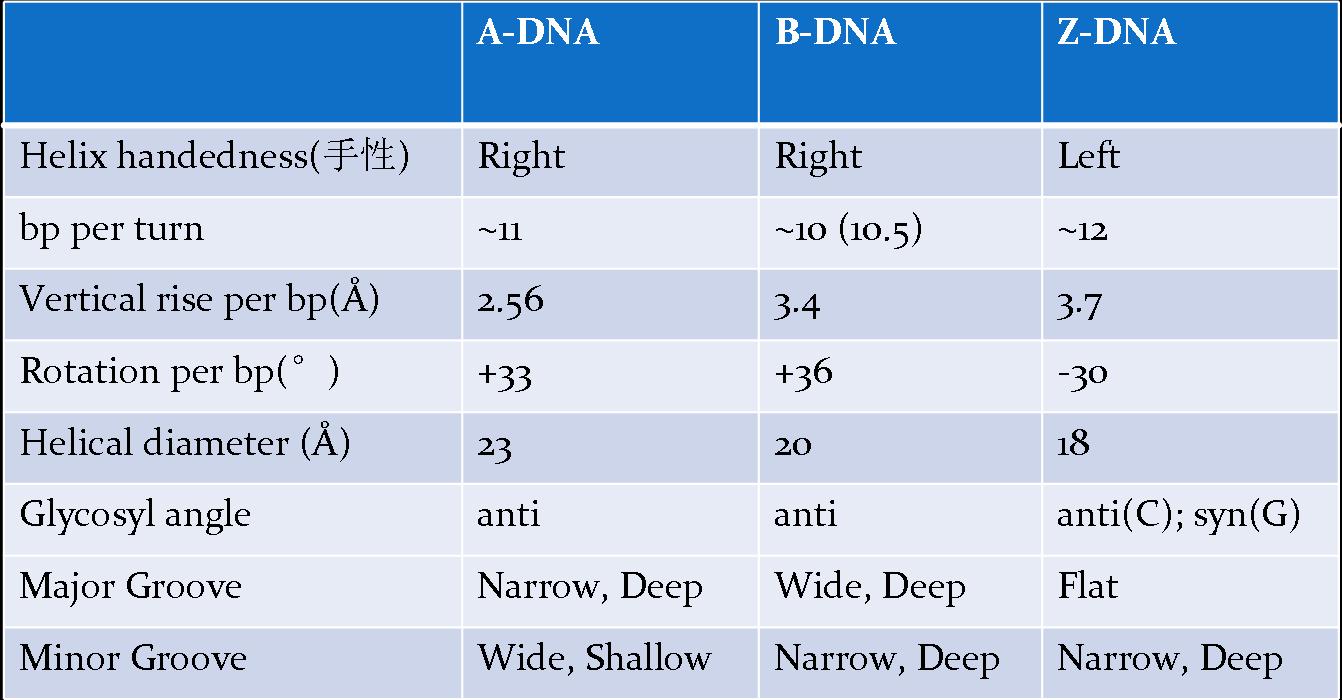
1. B-form:
- right-handed
- the structure identified by Watson and Crick,
- the most common form,
- believed to be the idealized form of the structure adopted by virtually all DNAin vivo (in the living body of a plant or animal), or, at physiological (characteristic of or appropriate to an organism’s healthy or normal functioning) pH and salt concentration.
characterized by:
- a helical repeat of 10bp/turn (although now it is known that ‘real’ B-DNA has a repeat closer to 10.5bp/turn);
- the presence of base pairs lying on the helix axis and almost perpendicular to it;
- having well-defined, deep major and minor grooves.
2. A-form:
- right handed
- adopted by DNA in vivo under unusual circumstances, (conversed from B-form in low moisture (<75%))
- presents in certain DNA-protein complexes
characterized by:
- a helical repeat of 11 bp/turn.
- the presence of base pairs tilted with respect to the helix axis, and actually lying off the axis.
- being the helix formed by RNA and DNA-RNA hybrids. (Similar to some RNA-DNA duplex or RNA-RNA duplex.)
3. Z-form:
- left-handed
- formed by stretches of alternating pyrimidine-purine sequence, e.g. GCGCGC, especially in negatively supercoiled DNA in high saline (盐) solution.
- not easily form even in DNA regions of repeating GCGCGC, since the boundaries between the left-handed Z-form and the surrounding B-form would be very unstale
characterized by:
- a zigzag (锯齿型) pattern where its name comes from
- 12 bp/turn
(1)
(2)
picture from Instant Notes in Molecular Biology (3rd Edtion)
====================================================
Some special structures of DNA
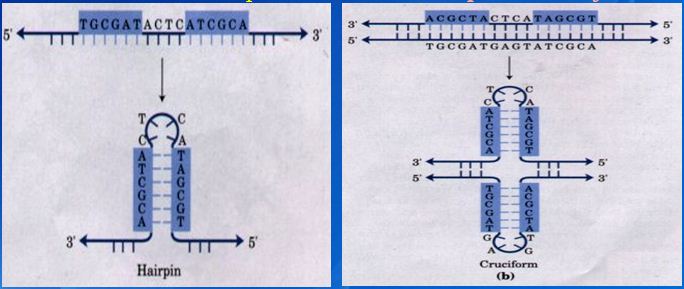
1. Inverted repeats and direct repeats
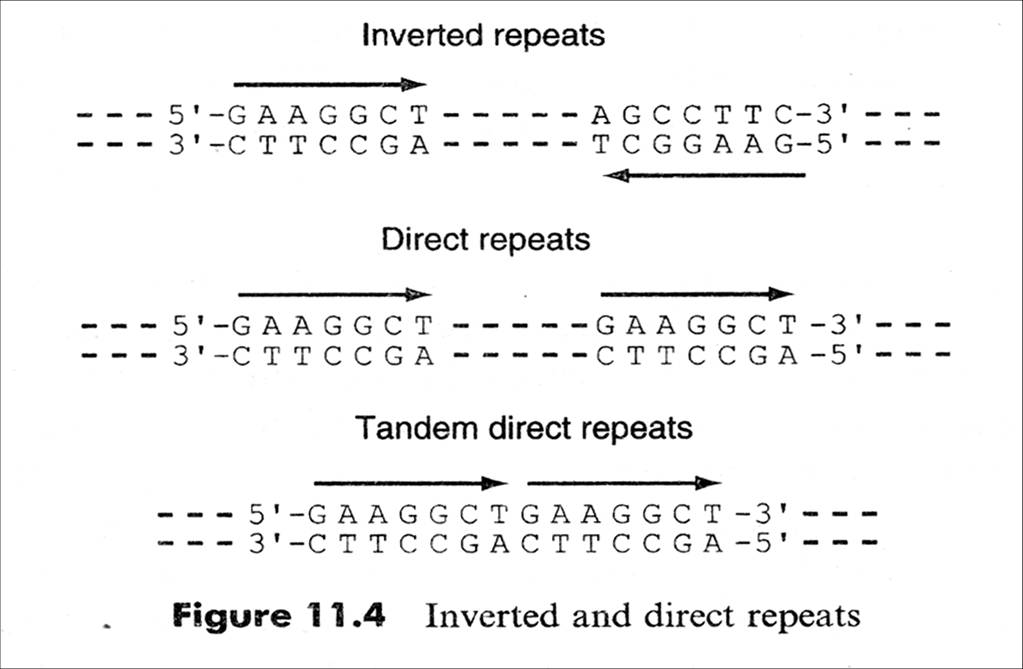
Inverted Repeat is functionally important as recognition sites on the DNA for the binding of a variety of proteins (e.g. restriction and modification enzymes)
Inverted Repeat is either discontinuous or continuous,
Continuous → palindromic structure (回文结构:inverted repetitions of base sequence over the two strands form a symmetric structure )
Discontinuous→ hairpin & cruciform (发卡、十字结构:self complementary within each of the strands )
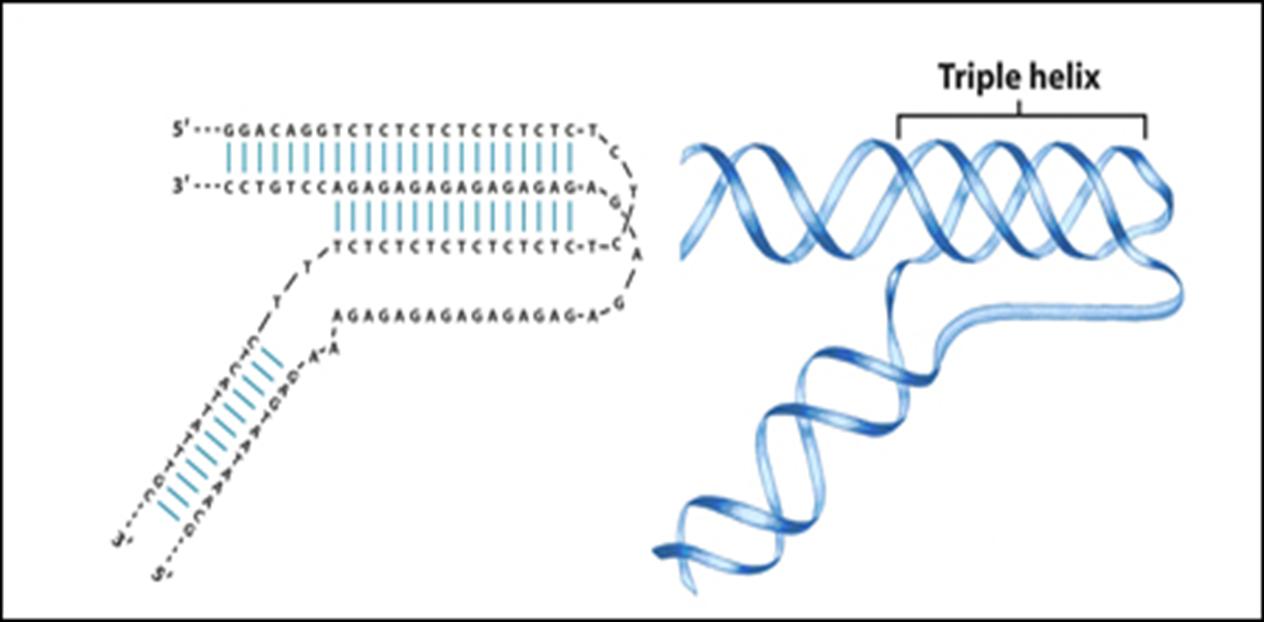
2. DNA triplex (The intramolecular triplex (H-DNA) as an example)
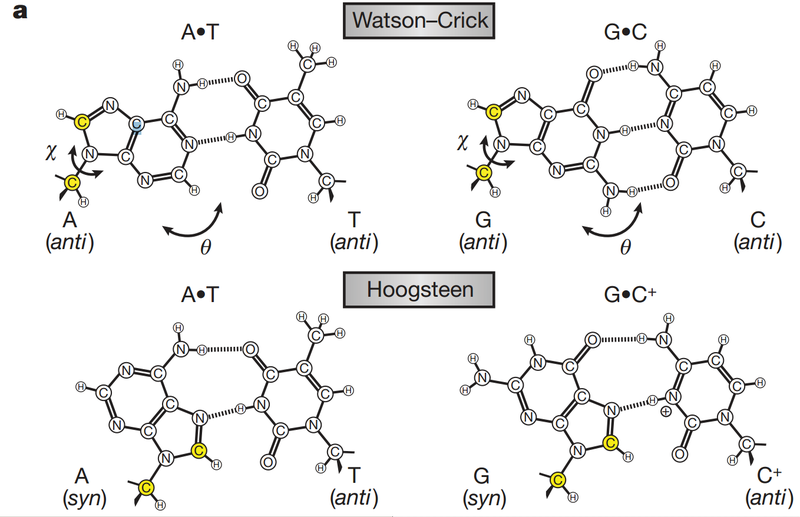
• AG-rich strand vs. CT-rich strand with Hoogsten hydrogen bond
(a quick glance at Hoodsten hydrogen bond:
• Mirror inverted repeat
• a barrier for different DNA and RNA polymerases, and so it is a negative regulator for gene expression.
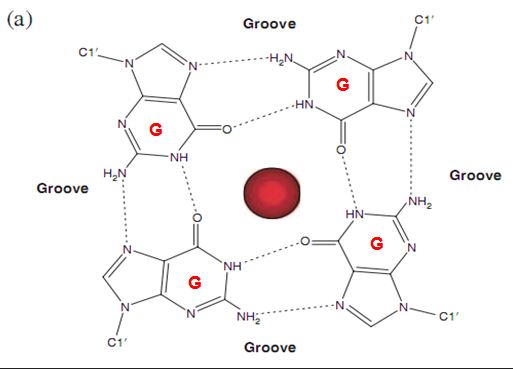
3. G-quadruplex structure